MS2 plot in mass_dataset
Xiaotao Shen PhD (https://www.shenxt.info/)
Created on 2020-03-28 and updated on 2022-09-19
Source:vignettes/ms2_plot_mass_dataset.Rmd
ms2_plot_mass_dataset.RmdData preparation
library(massdataset)
library(tidyverse)
library(metid)
ms1_data =
readr::read_csv(file.path(
system.file("ms1_peak", package = "metid"),
"ms1.peak.table.csv"
))
ms1_data = data.frame(ms1_data, sample1 = 1, sample2 = 2)
expression_data = ms1_data %>%
dplyr::select(-c(name:rt))
variable_info =
ms1_data %>%
dplyr::select(name:rt) %>%
dplyr::rename(variable_id = name)
sample_info =
data.frame(
sample_id = colnames(expression_data),
injection.order = c(1, 2),
class = c("Subject", "Subject"),
group = c("Subject", "Subject")
)
rownames(expression_data) = variable_info$variable_id
object = create_mass_dataset(
expression_data = expression_data,
sample_info = sample_info,
variable_info = variable_info
)
object
#> --------------------
#> massdataset version: 1.0.5
#> --------------------
#> 1.expression_data:[ 100 x 2 data.frame]
#> 2.sample_info:[ 2 x 4 data.frame]
#> 3.variable_info:[ 100 x 3 data.frame]
#> 4.sample_info_note:[ 4 x 2 data.frame]
#> 5.variable_info_note:[ 3 x 2 data.frame]
#> 6.ms2_data:[ 0 variables x 0 MS2 spectra]
#> --------------------
#> Processing information (extract_process_info())
#> create_mass_dataset ----------
#> Package Function.used Time
#> 1 massdataset create_mass_dataset() 2022-07-13 13:27:23
Add MS2 to mass_dataset object
path = "./example"
dir.create(path)
ms2_data <- system.file("ms2_data", package = "metid")
file.copy(
from = file.path(ms2_data, "QC1_MSMS_NCE25.mgf"),
to = path,
overwrite = TRUE,
recursive = TRUE
)
#> [1] TRUE
object =
massdataset::mutate_ms2(
object = object,
column = "rp",
polarity = "positive",
ms1.ms2.match.mz.tol = 10,
ms1.ms2.match.rt.tol = 30
)
#>
|
| | 0%
|
| | 1%
|
|= | 1%
|
|= | 2%
|
|== | 2%
|
|== | 3%
|
|== | 4%
|
|=== | 4%
|
|=== | 5%
|
|==== | 5%
|
|==== | 6%
|
|===== | 6%
|
|===== | 7%
|
|===== | 8%
|
|====== | 8%
|
|====== | 9%
|
|======= | 9%
|
|======= | 10%
|
|======= | 11%
|
|======== | 11%
|
|======== | 12%
|
|========= | 12%
|
|========= | 13%
|
|========= | 14%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 15%
|
|=========== | 16%
|
|============ | 16%
|
|============ | 17%
|
|============ | 18%
|
|============= | 18%
|
|============= | 19%
|
|============== | 19%
|
|============== | 20%
|
|============== | 21%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 22%
|
|================ | 23%
|
|================ | 24%
|
|================= | 24%
|
|================= | 25%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 26%
|
|=================== | 27%
|
|=================== | 28%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 29%
|
|===================== | 30%
|
|===================== | 31%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 32%
|
|======================= | 33%
|
|======================= | 34%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 35%
|
|========================= | 36%
|
|========================== | 36%
|
|========================== | 37%
|
|========================== | 38%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 39%
|
|============================ | 40%
|
|============================ | 41%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 42%
|
|============================== | 43%
|
|============================== | 44%
|
|=============================== | 44%
|
|=============================== | 45%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 46%
|
|================================= | 47%
|
|================================= | 48%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 49%
|
|=================================== | 50%
|
|=================================== | 51%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 52%
|
|===================================== | 53%
|
|===================================== | 54%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 55%
|
|======================================= | 56%
|
|======================================== | 56%
|
|======================================== | 57%
|
|======================================== | 58%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 59%
|
|========================================== | 60%
|
|========================================== | 61%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 62%
|
|============================================ | 63%
|
|============================================ | 64%
|
|============================================= | 64%
|
|============================================= | 65%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 66%
|
|=============================================== | 67%
|
|=============================================== | 68%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 69%
|
|================================================= | 70%
|
|================================================= | 71%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 72%
|
|=================================================== | 73%
|
|=================================================== | 74%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 76%
|
|====================================================== | 77%
|
|====================================================== | 78%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 79%
|
|======================================================== | 80%
|
|======================================================== | 81%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 82%
|
|========================================================== | 83%
|
|========================================================== | 84%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 86%
|
|============================================================= | 87%
|
|============================================================= | 88%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 89%
|
|=============================================================== | 90%
|
|=============================================================== | 91%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 92%
|
|================================================================= | 93%
|
|================================================================= | 94%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 96%
|
|==================================================================== | 97%
|
|==================================================================== | 98%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 99%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
| | 1%
|
|= | 1%
|
|= | 2%
|
|== | 2%
|
|== | 3%
|
|== | 4%
|
|=== | 4%
|
|=== | 5%
|
|==== | 5%
|
|==== | 6%
|
|===== | 6%
|
|===== | 7%
|
|===== | 8%
|
|====== | 8%
|
|====== | 9%
|
|======= | 9%
|
|======= | 10%
|
|======= | 11%
|
|======== | 11%
|
|======== | 12%
|
|========= | 12%
|
|========= | 13%
|
|========= | 14%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 15%
|
|=========== | 16%
|
|============ | 16%
|
|============ | 17%
|
|============ | 18%
|
|============= | 18%
|
|============= | 19%
|
|============== | 19%
|
|============== | 20%
|
|============== | 21%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 22%
|
|================ | 23%
|
|================ | 24%
|
|================= | 24%
|
|================= | 25%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 26%
|
|=================== | 27%
|
|=================== | 28%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 29%
|
|===================== | 30%
|
|===================== | 31%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 32%
|
|======================= | 33%
|
|======================= | 34%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 35%
|
|========================= | 36%
|
|========================== | 36%
|
|========================== | 37%
|
|========================== | 38%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 39%
|
|============================ | 40%
|
|============================ | 41%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 42%
|
|============================== | 43%
|
|============================== | 44%
|
|=============================== | 44%
|
|=============================== | 45%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 46%
|
|================================= | 47%
|
|================================= | 48%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 49%
|
|=================================== | 50%
|
|=================================== | 51%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 52%
|
|===================================== | 53%
|
|===================================== | 54%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 55%
|
|======================================= | 56%
|
|======================================== | 56%
|
|======================================== | 57%
|
|======================================== | 58%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 59%
|
|========================================== | 60%
|
|========================================== | 61%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 62%
|
|============================================ | 63%
|
|============================================ | 64%
|
|============================================= | 64%
|
|============================================= | 65%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 66%
|
|=============================================== | 67%
|
|=============================================== | 68%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 69%
|
|================================================= | 70%
|
|================================================= | 71%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 72%
|
|=================================================== | 73%
|
|=================================================== | 74%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 76%
|
|====================================================== | 77%
|
|====================================================== | 78%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 79%
|
|======================================================== | 80%
|
|======================================================== | 81%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 82%
|
|========================================================== | 83%
|
|========================================================== | 84%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 86%
|
|============================================================= | 87%
|
|============================================================= | 88%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 89%
|
|=============================================================== | 90%
|
|=============================================================== | 91%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 92%
|
|================================================================= | 93%
|
|================================================================= | 94%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 96%
|
|==================================================================== | 97%
|
|==================================================================== | 98%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 99%
|
|======================================================================| 100%
object
#>
[33m--------------------
#>
[39m
[32mmassdataset version: 1.0.5
#>
[39m
[33m--------------------
#>
[39m
[32m1.expression_data:
[39m[ 100 x 2 data.frame]
#>
[32m2.sample_info:
[39m[ 2 x 4 data.frame]
#>
[32m3.variable_info:
[39m[ 100 x 3 data.frame]
#>
[32m4.sample_info_note:
[39m[ 4 x 2 data.frame]
#>
[32m5.variable_info_note:
[39m[ 3 x 2 data.frame]
#>
[32m6.ms2_data:
[39m[ 25 variables x 24 MS2 spectra]
#>
[33m--------------------
#>
[39m
[32mProcessing information (extract_process_info())
#>
[39m
[32mcreate_mass_dataset ----------
#>
[39m Package Function.used Time
#> 1 massdataset create_mass_dataset() 2022-07-13 13:27:23
#>
[32mmutate_ms2 ----------
#>
[39m Package Function.used Time
#> 1 massdataset mutate_ms2() 2022-07-13 13:40:08
object@ms2_data
#> $`Mix_A_NCE25.mzXML;Mix_A_NCE25.mzXML;QC1_MSMS_NCE25.mgf`
#>
[33m--------------------
#>
[39m
[32mcolumn: rp
[39m
#>
[32mpolarity: positive
[39m
#>
[32mmz_tol: 10
[39m
#>
[32mrt_tol (second): 30
[39m
#>
[33m--------------------
#>
[39m
[32m25 variables:
#>
[39m
[32mpRPLC_603
[39m
[32mpRPLC_722
[39m
[32mpRPLC_778
[39m
[32mpRPLC_1046
[39m
[32mpRPLC_1112
[39m
[32m...
#>
[39m
[32m24 MS2 spectra.
#>
[39m
[32mmz162.112442157672rt37.9743312
[39m
[32mmz181.072050304971rt226.14144
[39m
[32mmz289.227264404297rt284.711172
[39m
[32mmz181.072050673093rt196.800648
[39m
[32mmz209.092155077047rt58.3735608
[39m
[32m...
#>
[39mIdentify metabolites according to MS2
data("snyder_database_rplc0.0.3", package = "metid")
object1 =
annotate_metabolites_mass_dataset(object = object,
database = snyder_database_rplc0.0.3)
#>
|
| | 0%
|
|=== | 4%
|
|====== | 8%
|
|======== | 12%
|
|=========== | 16%
|
|============== | 20%
|
|================= | 24%
|
|==================== | 28%
|
|====================== | 32%
|
|========================= | 36%
|
|============================ | 40%
|
|=============================== | 44%
|
|================================== | 48%
|
|==================================== | 52%
|
|======================================= | 56%
|
|========================================== | 60%
|
|============================================= | 64%
|
|================================================ | 68%
|
|================================================== | 72%
|
|===================================================== | 76%
|
|======================================================== | 80%
|
|=========================================================== | 84%
|
|============================================================== | 88%
|
|================================================================ | 92%
|
|=================================================================== | 96%
|
|======================================================================| 100%
head(object1@annotation_table)
#> variable_id ms2_files_id
#> 1 pRPLC_1046 Mix_A_NCE25.mzXML;Mix_A_NCE25.mzXML;QC1_MSMS_NCE25.mgf
#> 2 pRPLC_1046 Mix_A_NCE25.mzXML;Mix_A_NCE25.mzXML;QC1_MSMS_NCE25.mgf
#> 3 pRPLC_1046 Mix_A_NCE25.mzXML;Mix_A_NCE25.mzXML;QC1_MSMS_NCE25.mgf
#> 4 pRPLC_1112 Mix_A_NCE25.mzXML;Mix_A_NCE25.mzXML;QC1_MSMS_NCE25.mgf
#> 5 pRPLC_1307 Mix_A_NCE25.mzXML;Mix_A_NCE25.mzXML;QC1_MSMS_NCE25.mgf
#> 6 pRPLC_1860 Mix_A_NCE25.mzXML;Mix_A_NCE25.mzXML;QC1_MSMS_NCE25.mgf
#> ms2_spectrum_id Compound.name CAS.ID HMDB.ID KEGG.ID
#> 1 mz181.072050673093rt196.800648 Theophylline 611-59-6 HMDB01889 C07130
#> 2 mz181.072050673093rt196.800648 Paraxanthine 611-59-6 HMDB01860 C13747
#> 3 mz181.072050673093rt196.800648 Theophylline <NA> HMDB0001889 <NA>
#> 4 mz209.092155077047rt58.3735608 L-KYNURENINE <NA> <NA> <NA>
#> 5 mz314.232707486156rt400.268664 C10:1 AC <NA> HMDB13205 <NA>
#> 6 mz249.185015539689rt579.6807 C16:4 FA <NA> <NA> <NA>
#> Lab.ID Adduct mz.error mz.match.score RT.error RT.match.score CE
#> 1 RPLC_732 (M+H)+ 2.4391827 0.9952516 4.8006480 0.9872782 NCE25
#> 2 RPLC_456 (M+H)+ 1.9391827 0.9969962 6.4283218 0.9773041 NCE25
#> 3 RPLC_443 (M+H)+ 1.6891827 0.9977199 11.7251207 0.9264669 NCE25
#> 4 RPLC_252 (M+H)+ 0.1626926 0.9999788 0.4264392 0.9998990 NCE25
#> 5 RPLC_385 (M+H)+ 1.0812154 0.9990652 29.0070282 0.6265989 NCE25
#> 6 RPLC_529 (M+H)+ 1.0236508 0.9991621 15.6807000 0.8723170 NCE25
#> SS Total.score Database Level
#> 1 0.7523276 0.8717962 MS_0.0.2 1
#> 2 0.7523276 0.8697389 MS_0.0.2 1
#> 3 0.7523276 0.8572105 MS_0.0.2 1
#> 4 0.6469087 0.8234238 MS_0.0.2 1
#> 5 0.7931137 0.8029729 MS_0.0.2 1
#> 6 0.7315045 0.8336220 MS_0.0.2 1
head(extract_variable_info(object = object1))
#> variable_id mz rt Compound.name CAS.ID HMDB.ID KEGG.ID
#> 1 pRPLC_376 472.3032 772.906 <NA> <NA> <NA> <NA>
#> 2 pRPLC_391 466.3292 746.577 <NA> <NA> <NA> <NA>
#> 3 pRPLC_603 162.1125 33.746 L-Carnitine 541-15-1 HMDB00062 C00318
#> 4 pRPLC_629 181.0720 36.360 <NA> <NA> <NA> <NA>
#> 5 pRPLC_685 230.0701 158.205 <NA> <NA> <NA> <NA>
#> 6 pRPLC_722 181.0721 228.305 Theophylline <NA> HMDB0001889 <NA>
#> Lab.ID Adduct mz.error mz.match.score RT.error RT.match.score CE
#> 1 <NA> <NA> NA NA NA NA <NA>
#> 2 <NA> <NA> NA NA NA NA <NA>
#> 3 RPLC_406 (M+H)+ 1.667894 0.9977770 1.974331 0.9978368 NCE25
#> 4 <NA> <NA> NA NA NA NA <NA>
#> 5 <NA> <NA> NA NA NA NA <NA>
#> 6 RPLC_443 (M+H)+ 1.688262 0.9977224 17.615671 0.8416462 NCE25
#> SS Total.score Database Level
#> 1 NA NA <NA> NA
#> 2 NA NA <NA> NA
#> 3 0.6048288 0.8013178 MS_0.0.2 1
#> 4 NA NA <NA> NA
#> 5 NA NA <NA> NA
#> 6 0.6071017 0.7633930 MS_0.0.2 1MS2 plot
ms2_plot_mass_dataset(object = object1, variable_index = 1, database = snyder_database_rplc0.0.3)
#> NULLms2_plot_mass_dataset(object = object1, variable_index = 3, database = snyder_database_rplc0.0.3)
#> $pRPLC_603_1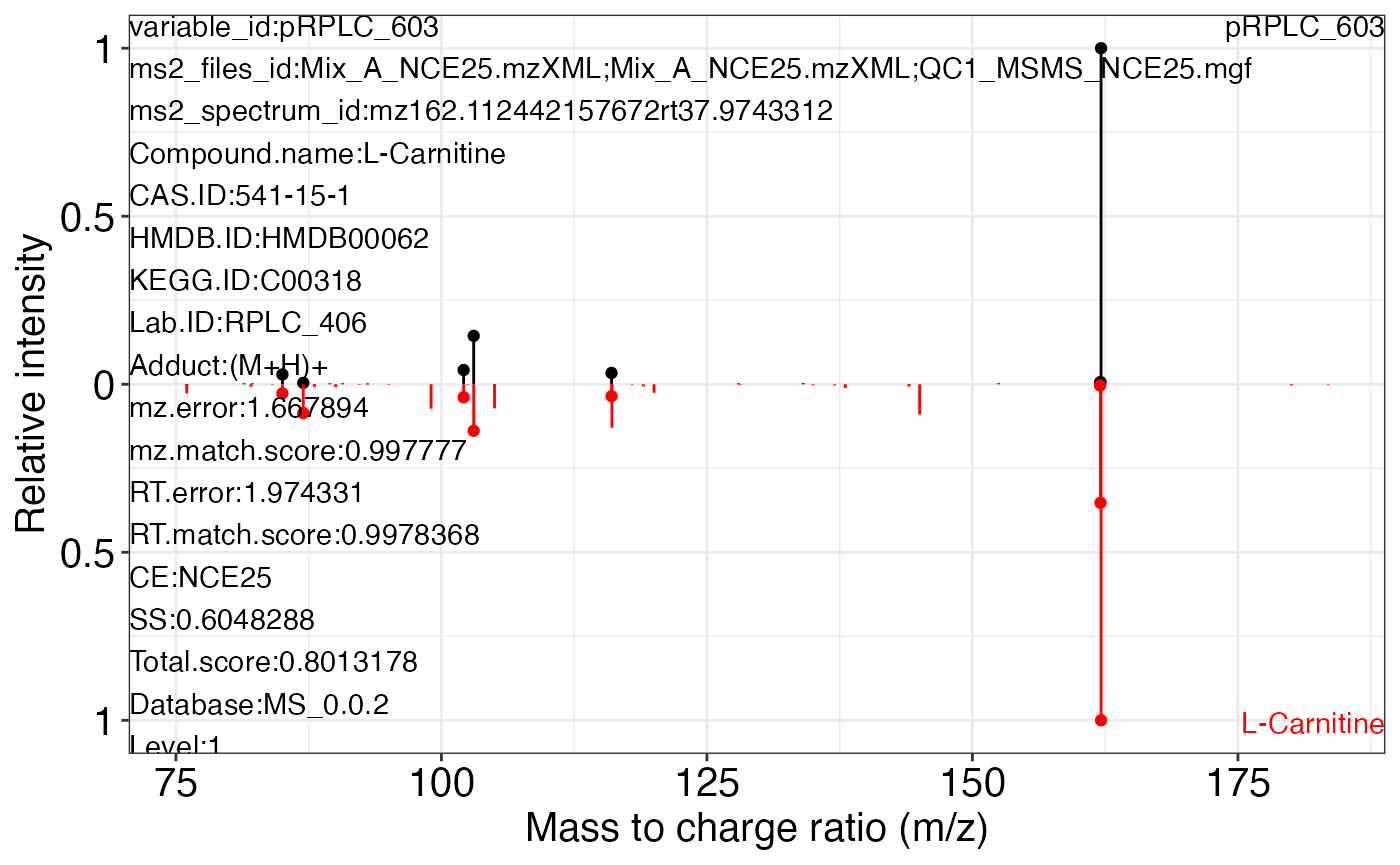
Session information
sessionInfo()
#> R version 4.2.1 (2022-06-23)
#> Platform: x86_64-apple-darwin17.0 (64-bit)
#> Running under: macOS Big Sur ... 10.16
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] metid_1.2.16 forcats_0.5.1.9000 stringr_1.4.0 dplyr_1.0.9
#> [5] purrr_0.3.4 readr_2.1.2 tidyr_1.2.0 tibble_3.1.7
#> [9] tidyverse_1.3.1 tinytools_0.9.1 ggplot2_3.3.6 magrittr_2.0.3
#> [13] masstools_0.99.13 massdataset_1.0.5
#>
#> loaded via a namespace (and not attached):
#> [1] readxl_1.4.0 backports_1.4.1
#> [3] circlize_0.4.15 systemfonts_1.0.4
#> [5] plyr_1.8.7 lazyeval_0.2.2
#> [7] listenv_0.8.0 BiocParallel_1.30.3
#> [9] GenomeInfoDb_1.32.2 Rdisop_1.56.0
#> [11] digest_0.6.29 foreach_1.5.2
#> [13] yulab.utils_0.0.5 htmltools_0.5.2
#> [15] fansi_1.0.3 memoise_2.0.1
#> [17] cluster_2.1.3 doParallel_1.0.17
#> [19] tzdb_0.3.0 openxlsx_4.2.5
#> [21] limma_3.52.2 globals_0.15.1
#> [23] ComplexHeatmap_2.12.0 modelr_0.1.8
#> [25] matrixStats_0.62.0 vroom_1.5.7
#> [27] pkgdown_2.0.5 prettyunits_1.1.1
#> [29] colorspace_2.0-3 rvest_1.0.2
#> [31] textshaping_0.3.6 haven_2.5.0
#> [33] xfun_0.31 crayon_1.5.1
#> [35] RCurl_1.98-1.7 jsonlite_1.8.0
#> [37] impute_1.70.0 iterators_1.0.14
#> [39] glue_1.6.2 gtable_0.3.0
#> [41] zlibbioc_1.42.0 XVector_0.36.0
#> [43] GetoptLong_1.0.5 DelayedArray_0.22.0
#> [45] shape_1.4.6 BiocGenerics_0.42.0
#> [47] scales_1.2.0 vsn_3.64.0
#> [49] DBI_1.1.3 Rcpp_1.0.8.3
#> [51] mzR_2.30.0 progress_1.2.2
#> [53] viridisLite_0.4.0 clue_0.3-61
#> [55] gridGraphics_0.5-1 bit_4.0.4
#> [57] preprocessCore_1.58.0 stats4_4.2.1
#> [59] MsCoreUtils_1.8.0 htmlwidgets_1.5.4
#> [61] httr_1.4.3 RColorBrewer_1.1-3
#> [63] ellipsis_0.3.2 farver_2.1.1
#> [65] pkgconfig_2.0.3 XML_3.99-0.10
#> [67] sass_0.4.1 dbplyr_2.2.1
#> [69] utf8_1.2.2 labeling_0.4.2
#> [71] ggplotify_0.1.0 tidyselect_1.1.2
#> [73] rlang_1.0.3 munsell_0.5.0
#> [75] cellranger_1.1.0 tools_4.2.1
#> [77] cachem_1.0.6 cli_3.3.0
#> [79] generics_0.1.3 broom_1.0.0
#> [81] evaluate_0.15 fastmap_1.1.0
#> [83] mzID_1.34.0 yaml_2.3.5
#> [85] ragg_1.2.2 bit64_4.0.5
#> [87] knitr_1.39 fs_1.5.2
#> [89] zip_2.2.0 ncdf4_1.19
#> [91] future_1.26.1 pbapply_1.5-0
#> [93] xml2_1.3.3 compiler_4.2.1
#> [95] rstudioapi_0.13 plotly_4.10.0
#> [97] png_0.1-7 affyio_1.66.0
#> [99] reprex_2.0.1 bslib_0.3.1
#> [101] stringi_1.7.6 highr_0.9
#> [103] desc_1.4.1 MSnbase_2.22.0
#> [105] lattice_0.20-45 ProtGenerics_1.28.0
#> [107] Matrix_1.4-1 ggsci_2.9
#> [109] vctrs_0.4.1 furrr_0.3.0
#> [111] pillar_1.7.0 lifecycle_1.0.1
#> [113] BiocManager_1.30.18 jquerylib_0.1.4
#> [115] MALDIquant_1.21 GlobalOptions_0.1.2
#> [117] data.table_1.14.2 bitops_1.0-7
#> [119] GenomicRanges_1.48.0 R6_2.5.1
#> [121] pcaMethods_1.88.0 affy_1.74.0
#> [123] parallelly_1.32.0 IRanges_2.30.0
#> [125] codetools_0.2-18 MASS_7.3-57
#> [127] assertthat_0.2.1 SummarizedExperiment_1.26.1
#> [129] rprojroot_2.0.3 rjson_0.2.21
#> [131] withr_2.5.0 S4Vectors_0.34.0
#> [133] GenomeInfoDbData_1.2.8 parallel_4.2.1
#> [135] hms_1.1.1 grid_4.2.1
#> [137] rmarkdown_2.14 MatrixGenerics_1.8.1
#> [139] Biobase_2.56.0 lubridate_1.8.0